Abstract
Introduction: Acute graft-versus-host disease (aGVHD) is a severe complication of allogeneic hematopoietic cell transplant (allo-HCT) and a common cause of non-relapse mortality in patients receiving allo-HCT. In aGVHD, alloreactive donor T cells attack host tissues, ultimately leading to end-organ damage primarily of the liver, skin, and gastrointestinal tract. Using murine models, prior work from our lab has shown that microRNA-155 (miR-155) is necessary for aGVHD development. Moreover, we have reported that transplanting T cells from miR-155 knockout mice completely prevent aGVHD. Importantly, transplanting miR-155 knockout T cells retains a graft-versus-leukemia (GVL) response, required for the prevention of relapse. We have also demonstrated that targeting miR-155 in vivo with antisense oligonucleotides against miR-155 (antimiR-155) leads to lower aGVHD severity; however, the efficiency of targeting miR-155 was low, and the responses were not robust despite using antimiR-155 then undergoing clinical trials and optimizing a dose and schedule in mice. So, there is an unmet need to target miR-155 using different strategies. Methods: We used CRISPR/Cas9 genome editing to target miR-155 host gene (MIR155HG) in primary human donor T cells. We designed different pairs of single guide RNAs (sgRNAs), targeting the promoter region (Pr) or exon 3 (Ex3) of MIR155HG, while a non-targeting (NT) guide was used as control. Knockout was confirmed at DNA level by genomic PCR (gPCR) and at RNA level by assessing the expression of mature miR-155 transcript by qRT-PCR. CRISPR/Cas9 on target efficiency was assessed by digital droplet PCR (ddPCR) while off-target effects were analyzed by Whole Genome Sequencing (WGS) followed by CHURCHILL genome analysis. To evaluate the functionality of edited human donor T cells, we performed xenogeneic aGVHD transplants with MIR155HG edited or control human donor T cells injected into NSG mice. To assess the ability of edited T cells to maintain GVL response, we are currently performing an in vivo GVL study using leukemic (MOLM-13) cells alone or MOLM-13 along with MIR155HG edited or control human donor T cells injected into NSG mice.
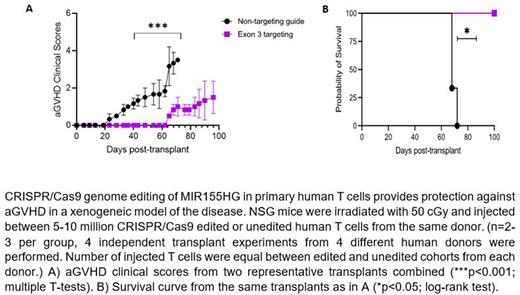
Results: Predicted promoter and exon3 region deletions were confirmed by gPCR, with the size difference observed in the PCR products obtained from non-targeting (NT) control vs. Ex3 (418bp vs. 243bp) or Pr (551bp vs. 284bp) edited genomic DNA. qRT-PCR showed that the expression of mature miR-155 transcript at 72 hrs was significantly reduced relative to non-targeting (NT) control guide (fold change- Ex3 vs. NT control = 0.33 vs. 1, p<0.0001; Pr vs. NT control = 0.48 vs. 1, p<0.01). The reduced miR-155 transcript level remained substantially sustained in Ex3 cells after 7 days of expansion; hence only exon3 targeting guides were chosen for further research. ddPCR showed a mean 49.05% editing (n=4) using the Ex3 targeting guides (p<0.01). WGS and CHURCHILL analysis performed on DNA from unedited and Ex3 samples from the same human donor identified 6 off-target genes (HDAC7, KPNA4, MCC, OLFML2A, HRNR and IGFN1), out of which only HDAC7 and KPNA4 were found to be expressed in T cells, as assessed by qRT-PCR performed in unstimulated versus CD3/CD28 stimulated T cells. There was no significant change in gene and protein expression of HDAC7 (Ex3 vs. NT control = 1.19 vs. 1, ns) and KPNA4 (0.86 vs. 1, ns) between the NT control and Ex3 edited samples among three human donors. In xenogeneic aGVHD transplant study, we observed that the recipients of MIR155HG deleted human donor T cells display significantly lower aGVHD clinical scores (p<0.001, Figure A), significantly improved survival compared to mice receiving control T cells (Figure B; 68-days in NT control vs. median survival not reached in Ex3, p<0.05), and significantly lower average histopathological scores, respectively in Ex3 vs. NT control in liver (3.25 vs. 13.4, respectively; p<0.05) and skin (1.16 vs. 2.50, respectively; p<0.05).
Conclusion: CRISPR/Cas9 mediated deletion of MIR155HG in donor T cells provides protection against lethal aGVHD in the xenogeneic model.
Significance: Our studies support the development of genome editing of MIR155HG in primary human donor T cells as a novel and feasible strategy to prevent aGVHD.
Disclosures
Braunreiter:Sarepta Therapeutics: Current Employment, Current holder of stock options in a privately-held company. Naeimi Kararoudi:Kids/Sanofi: Patents & Royalties. Malik:Aruvant Sciences: Consultancy, Patents & Royalties, Research Funding. Choe:Abbvie: Other: independent endpoint review committee for trial. Ranganathan:Plexxikon Inc: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.